Agarose gel electrophoresis separates dna fragments by ____. We present detailed explanations to you under the headings. Here are the details.
Agarose gel electrophoresis separates dna fragments by ____.
Agarose gel electrophoresis separates dna fragments by ____.
- Question:
Gel electrophoresis separates DNA fragments according to their _____.
- (a) base sequence
- (b) size
- (c) percentage of labelled nucleotides
- (d) electrical charge.
The correct answer to this question is option b. DNA in agarose or acrylamide gels is separated by size as all DNA molecules are equally (negative) charged, i.e. if the DNA fragments in a mixture are different, they only do so by sequence and size.
Differences in sequence are used to analyze DNA by distinguishing their hybridization abilities (detection by sequence similarity), while physical separation of DNA molecules is best achieved by exploiting their indiscriminate differences in size. During gel electrophoresis, smaller pieces work much faster than larger pieces. Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.
In this article, we have published a DNA migration technique that can be performed in the classroom with agarose gel electrophoresis. We will also indicate how to highlight migratory bands that are invisible to the naked eye. It recommends using online DNA sequence databases to estimate the number of DNA segments that need to be obtained based on the restriction enzymes involved and compare this with the results of electrophoresis performed in the classroom.
1. Introduction
Agarose gel electrophoresis separates dna fragments by ____. Detailed continues. Numerous scientific and technological applications involving DNA study are fed by different techniques. Of course, electrophoresis on agarose or polyacrylamide gel is one of the most used. This is because it makes it possible to separate molecules based on their size. This is an essential prerequisite in many applications for identifying a gene, especially identifying the pieces of DNA cut by enzymes.
DNA electrophoresis on agarose gel presents little technical difficulty. It can be easily used in practical work both at university and high school to introduce students to this basic technique. The analysis of results, combined with the various computer tools (word processing, spreadsheet, image acquisition and processing software) commonly used in our time, can lead to significant advances that allow for a better understanding of the scientific and practical relevance of DNA technologies. The different possibilities for using these various tools are outlined below.
2. Practice of DNA agarose gel electrophoresis
Agarose gel electrophoresis separates dna fragments by ____. Detailed continues. Practice of DNA agarose gel electrophoresis, all details and answers below.
2.1. Agarose gel
- Mix TBE buffer and agarose at the rate of 0.8 g of agarose for 100 mL of buffer (the proportion of agarose depends on the size of the DNA molecules to be separated).
- Melt the agarose in the microwave oven, watching to avoid splashing, or in a bain-marie. Shake occasionally to homogenize the mixture.
- Allow to cool until the bottle can be picked up with bare hands (approximately 60°C).
- Place the gaskets supplied with the tank to close the gel support and position the comb 1 mm from the bottom and about 1 cm from the end of the support. Adjust the level so that the gel holder is horizontal. Some combs are adjustable with a screw: there is then a wedge to adjust their height. Others are pre-adapted to the support and do not require a wedge.
- Slowly pour the gel 3 to 5 mm thick, ensuring that it surrounds the teeth of the comb.
- Leave to cool, remove the comb and seals. The gel is ready for sample loading.
NB: Gel can be prepared in advance and kept solid in the refrigerator in a glass bottle (2/3 full maximum). At the time of use, remelt the gel in the microwave oven (open the cap and monitor to avoid splashing) or in a boiling water bath (open the cap). Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.
2.2. Preparation and deposit of samples
Agarose gel electrophoresis separates dna fragments by ____. The easiest way to get used to this technique without difficulty is, of course, to buy DNA samples from the market, either whole or digested with restriction enzymes. Suppliers offer a variety of enzyme-digested and non-enzyme-digested DNA from different sources.
DNA samples can be obtained in solution in TE buffer (Tris EDTA) or lyophilized. In this case, they are redissolved in TE buffer. DNA solutions can be stored indefinitely at -20°C.
DNA samples are generally distributed by suppliers with loading dye, which must be mixed with the samples before storage, following the proportions specified by the manufacturer. It’s important to have a precision pipette for the rest of the operations. Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.
Protocol
- Place the DNA samples for 3 min at 60-65°C.
- Transfer them to ice for 2-3 min.These two operations are intended to prevent the ligation of fragments possibly having cohesive ends.
For filling 10-12 mm wide wells well suited for educational use, a proportion of 5 µL of loading dye for 10 to 15 µL of DNA is suitable (so as to obtain 1 to 4 µg of DNA per well). - Mix the loading dye and the DNA on a piece of parafilm and withdraw the mixture with a micropipette adjusted to the appropriate volume, changing tips with each withdrawal.
- Fill the wells being careful not to tear the bottom of the gel with the tip of the pipette.
- Place the support with the loaded gel in the electrophoresis tank, positioning the wells on the cathode side (black pole).
- Fill the tank with TBE buffer (reusable several times) by pouring delicately and very slowly when the gel begins to be covered to prevent DNA from leaking into the buffer.
- Close the tank, connect the wires and turn on the power.
- Allow to migrate until the loading dye comes close to the edge of the gel (approximately 55 min at 100 V for an 80 mm gel in a minicuvette).
- Cut the power supply, unplug the connections and recover the gel in its holder.
Be careful when transporting: the gel slips very easily from its support. Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.

2.3. Revealing DNA bands
Please note that the gel must be handled with care as it is very fragile. Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.
- Cover the gel with the dye.
- Leave to act for 1 hour to overnight then empty the dye (which can be reused). DNA bands appear blue.
The gels are observable at this stage but you may want to discolor the background to improve the quality of the photographs. - Rinse in distilled water for a few hours, changing the water occasionally.
- Photograph the gel with a digital camera or scanner.
The pictures below, taken with a scanner, show the results obtained for two sizes of agarose gels.
You should know that the contrast of images is enhanced by digital image processing to avoid loss of information caused by shooting. / Agarose gel electrophoresis separates dna fragments by ____.
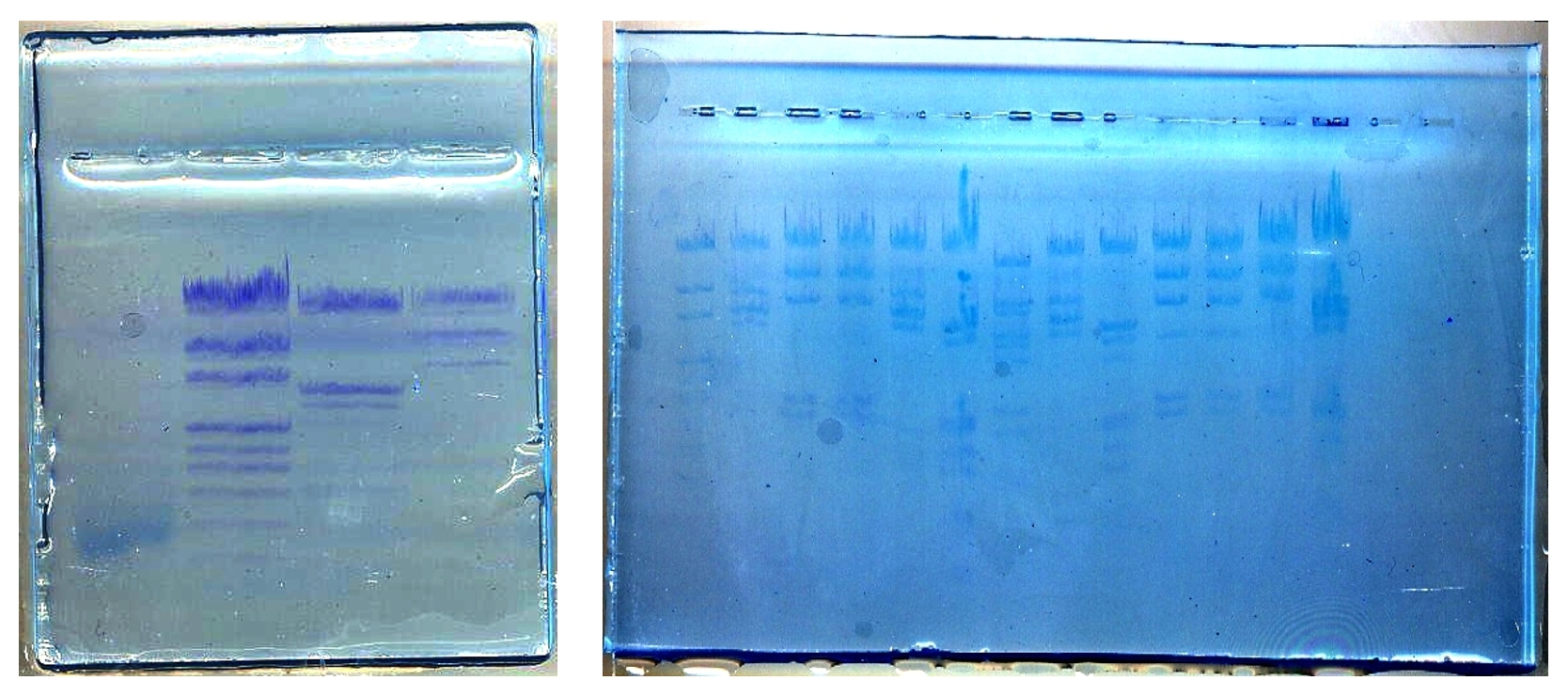
Minigel tracks:
| 1 | 2 | 3 | 4 |
| lentil genomic DNA | Size markers | Lambda phage DNA digested with EcoRI and HindIII | Lambda phage DNA digested with HindIII |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| pUC Mix (P) | EcoRI (P) | Hind III (P) | Hind III (S) | EcoRI (P) | EcoRI + HindIII (P) | BstII (S) | EcoRI (S) | EcoRI + HindIII (P) | Hind III (P) | Hind III (P) | Hind III (P) | EcoRI + HindIII (P) |
Wells 2 to 13: lambda phage DNA digested with various restriction enzymes. DNA Origin: Promega: P; Sigma:S / Agarose gel electrophoresis separates dna fragments by ____.
3. Image processing and detection of bands invisible to the naked eye
Image processing and detection of bands invisible to the naked eye, all details and answers are below.
3.1. Simple Processing
In particular, the visibility of bands marked less with paint can be further improved by digitally processing the images with image processing software (PhotoEditor, Paint Shop Pro, Photoshop, etc.). Adjustments can be made on brightness, contrast and relative intensity of colors. You can also have a generally more readable negative image like the one are below. / Agarose gel electrophoresis separates dna fragments by ____.
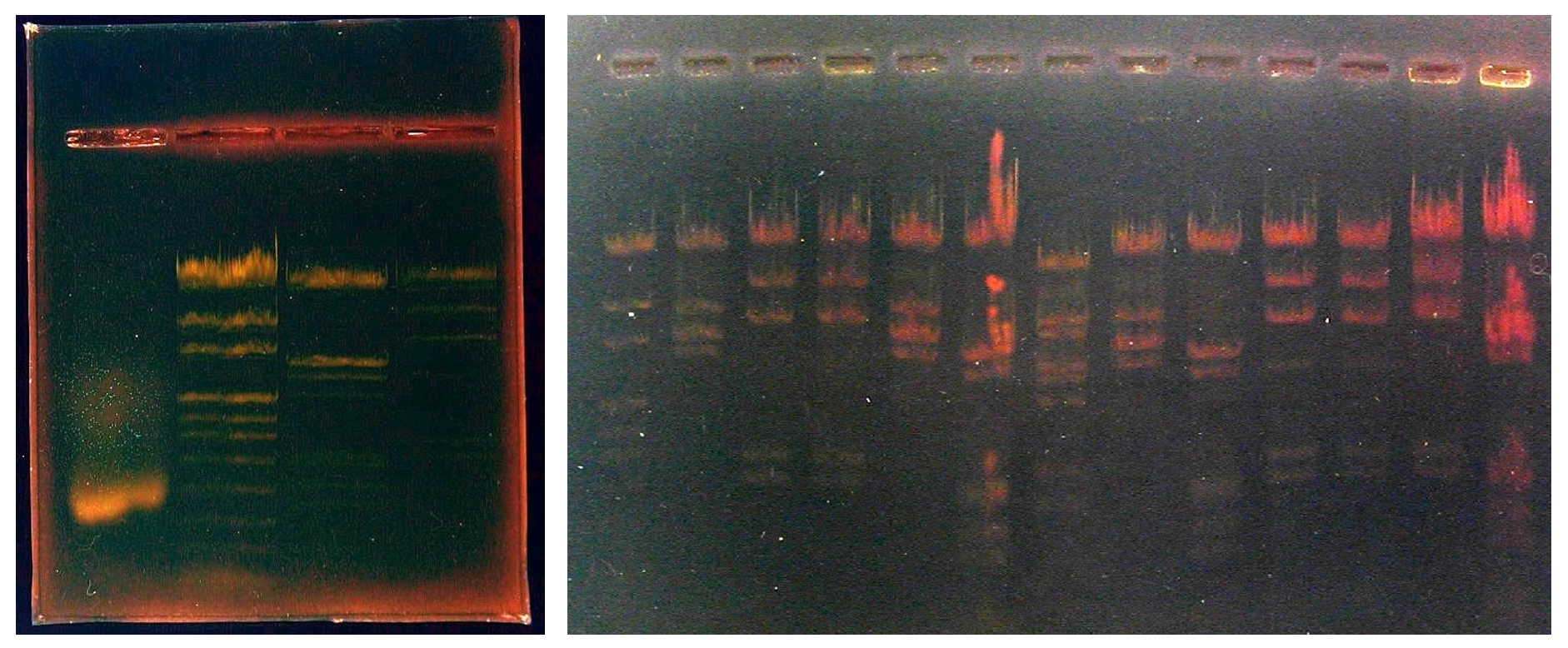
3.2. Detection of invisible bands by digital processing
Agarose gel electrophoresis separates dna fragments by ____. Detailed continues. Digital image processing software Digispec has the ability to measure the light intensity of each pixel of the image and its tricolor component. The graphical representation of each of these graphs, using a spreadsheet software such as Excel, plays a role in detecting bands invisible to the naked eye by describing the intensity changes on the curves. The figure below represents the graphs obtained with band 2 of the above minigel (length markers, pUCMix).
Another software, Mesurim, developed by JF Madre and freely downloadable from the site of the IFE (French Institute of Education) also allows this type of processing. Refer to the help sections available on this site for its use. / Agarose gel electrophoresis separates dna fragments by ____.
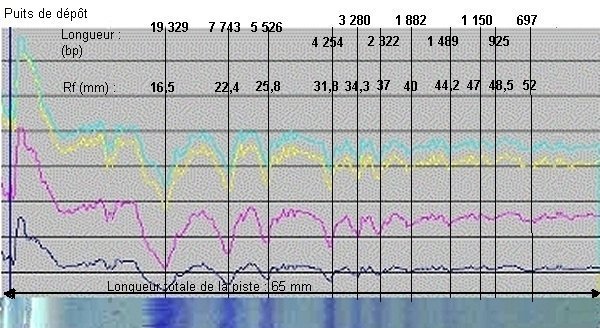
Note that only 9 bands are visible to the naked eye in this lane 1, but simultaneous variations of the four densitometric plots allow two additional bands (925 and 697 base pairs, respectively) to be identified. Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.
Migration distances will be measured by defining the bands. However, to do this, the resulting image will be displayed in image processing software (PhotoEditor, Paint Shop Pro, Photoshop, etc.) and the distance from each band to the well will be measured directly with the mouse; The software displays the number of corresponding pixels.
The resulting values are indicated in the graph above (Rf in mm). Knowing the size in base pairs of the DNA fragments delivered by the supplier, it is possible to construct the curve expressing the migration distance as a function of the size of the DNA fragments.
4. Construction of the curve Migration distance = f (DNA size)
Agarose gel electrophoresis separates dna fragments by ____. Detailed continues. Generating the curve requires, on the one hand, information given by the supplier about the size of the various constraints in the mix, and on the other hand, previous measurements on the image to determine the distance.
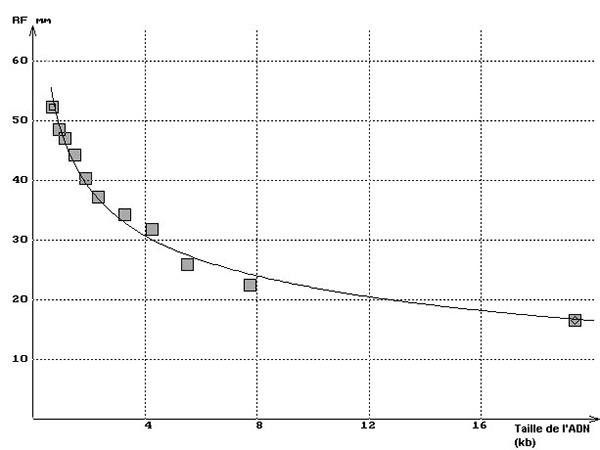
Any spreadsheet can be used to enter data (size of segments on abscissa, transition distances on ordinate) and generate the corresponding graph (Rf = a(1/log length) + b) that creates the standard curve. You can determine the size of the unknown fragments from their migration distances. Also, the nature of the curve plays an important role in checking the good adequacy of the results obtained with the size of the restriction pieces specified by the supplier.
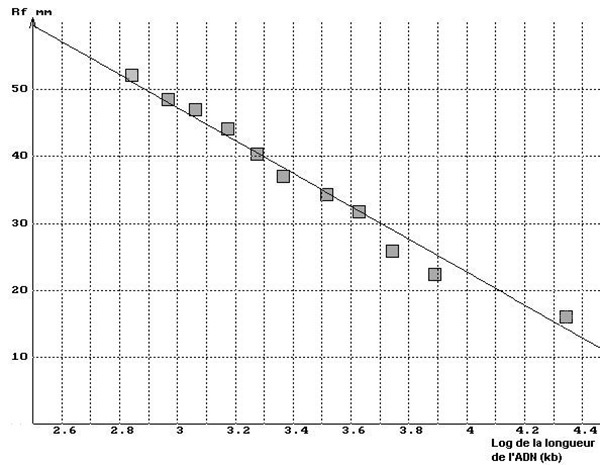
Agarose gel electrophoresis separates dna fragments by ____. In the graph below on the left, you’ll see the results with strip 2 of the minigel above and the theoretical curve superimposed. The graph will also show how the DNA segments on the apse can be linearized using a logarithm of size or a semi-logarithmic scale, as shown in the right graph below.
5. Determination of the length of DNA fragments of unknown size
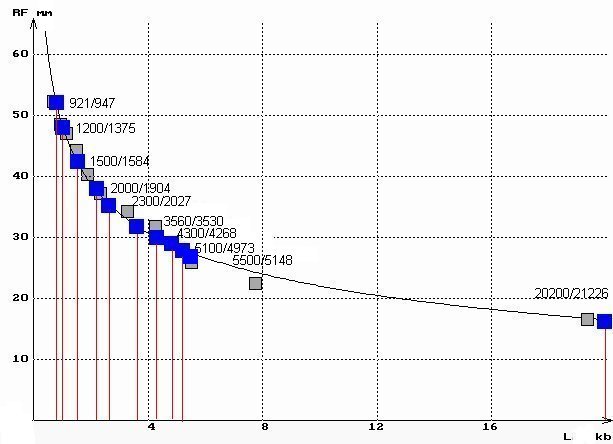
Agarose gel electrophoresis separates dna fragments by ____. Detailed continues. The graphs above can be used to determine the size of unknown DNA fragments separated by electrophoresis. For this, the migration distance of bands of unknown size is measured on the graph. We identified bands invisible to the naked eye as outlined above by analysis of the lane image, and then proceeded in this way for lane 3 of the above minigel. (phage lambda DNA was fully digested with EcoRI and HindIII). The figure below shows the results of this process.
6. Location of restriction sites and prediction of results
Agarose gel electrophoresis separates dna fragments by ____. DNA sequences stored in international databases (see end of article) are accessible directly from web servers where they can be easily downloaded. For the examples given here, the DNA sequence of bacteriophage lambda can be downloaded directly from this page (in html format or in .txt text format that can be used directly with a word processor) by clicking on the corresponding link. Agarose gel electrophoresis separates dna fragments by ____. Detailed continues.
The lambda phage genome comprises 48,502 base pairs and has a molecular mass of 31.5 x 10 6 daltons.
- Sequence of the phage lambda genome (to save the file, ask to save the target of the link).
Once the sequence has been loaded, one can undertake to locate the restriction sites in the genome of the lambda phage whose DNA was used to carry out the electrophoresis.
The EcoRI enzyme recognizes the sequence:
- ..GAATTC..
- ..CTTAAG..
The HindIII enzyme recognizes the sequence:
- ..AAGCTT..
- ..TTCGAA..
Agarose gel electrophoresis separates dna fragments by ____. The search function of any word processor or browser can locate restriction sites if it is told the sequence to be found. Note that the sequence must not contain spaces between the nucleotides so that the “search” function can find all the restriction sites.
Under these conditions, the search for the GAATTC sequence leads to the identification of five restriction sites located respectively at 21226, 26104, 31747, 39168, 44972 generating six fragments. Similarly, the search for the AAGCTT sequence leads to the identification of six restriction sites located respectively at 23130, 25157, 27479, 36895, 37459, 44141, generating seven fragments.
7. Practical information
Agarose gel electrophoresis separates dna fragments by ____. Practical Information, all details below.
7.1. Solutions
- TBE buffer (Tris borate EDTA) pH 8.3
For 1000 mL of distilled water:
Tris.HCl [tris(hydroxymethyl)aminomethane] 90 mmol.L-1: 10.89 g
Boric acid 90 mmol.L-1: 5 .56 g
EDTA 2 mmol.L-1: 0.74 g (disodium salt of ethylenediaminetetraacetic acid)
- TE buffer (Tris EDTA) pH 7.6
For 100 mL of distilled water:
– Tris.HCl [tris(hydroxymethyl)aminomethane] 1 mol.L-1: 12.1 g
– EDTA (disodium salt of ethylene acid tetraacetic diamine) 0.1 mol.L-1: 3.72 g
- Filling dye pH 8
For 10 mL of distilled water:
– Bromophenol blue 3 mmol.L-1: 0.2 g
– Sucrose 1.5 mol.L-1: 5.1 g
– Tris. HCl 10 mmol.L-1: 0.01 g
- Staining solution for gels
– Stock solution (100 times concentrated) to be stored in the refrigerator: Dissolve 10 mg of Nile blue in 5 mL of distilled water.
– Staining solution: 1 mL of the stock solution for 100 mL of distilled water.
Depending on the size of the staining tank, prepare 200 to 300 mL of the staining solution.
7.2. Supply
- Electrophoresis tanks: Pierron , Bio-Rad , Bioblock
- Chemicals: SIGMA . L’Isle d’Abeau Chesnes BP 701 – 38297 Saint Quentin Fallavier
- DNA: EUROMEDEX. BP 80 – 67460 Souffelweyersheim
- DNA sequences: EMBL , Genbank (For phage lambda genome, accession numbers: M17233, M24325, V00636)
Agarose gel electrophoresis separates dna fragments by ____. Video are below.
- What is the domain of the function mc007-1.jpg? mc007-2.jpg mc007-3.jpg mc007-4.jpg mc007-5.jpg.
- What İs The Domain Of The Function Shown İn The Mapping?
- How Many Resonance Structures Exist For The Formate İon Hco2
- Division Algorithm For Polynomials Proof
- Methanol Ethanol And N-propanol Are Three Common Alcohols